Providing food, fuel, feed, and many other important resources for human beings, crops fulfill the rapidly increasing global food demand and thus save our lives and greatly contribute to society (Tilman et al., 2011). The world’s population is overgrowing and there’s no sign of populations coming into the control mark. The decrease in land and pure water supply are the result of this phenomenon. In addition, climate change and environmental hazards such as frost, drought, flood, and earthquake are making the survival of humanity more difficult. A sustainable agricultural method and crop improvement is a must in this situation (El-Mounadi et al., 2020). Genetic engineering come forward with its diverse technological breakthroughs to improve the traits of agriculturally important crop plants. The development of insect or pest-resistant, herbicide-tolerant, virus-resistant, drought and frost-tolerant crops with higher yields and improved nutritional values is becoming possible via the implementation of genetic engineering tools (Eş et al., 2019). The discovery of the functions of many crop genes and the sequencing of crop genomes have been done with the rapid development of the sequencing process. Now, this genomic information of crops is widely used to improve and cultivate better crops with important agronomic traits and higher yields (Jeon et al., 2000). The genomic study of crops facilitates scientists to understand the significance of certain genes for certain characteristics of crops. A wide array of genome editing tools such as CRISPR/Cas9, ZFNs, TALENS, EMNs (Engineered endonucleases or meganucleases), were introduced to improve conventional breeding and agricultural techniques. These genome editing technologies insert or delete specific genes or induce mutations in target genes to produce available crops with improved characteristics (Nagamine and Ezura, 2022; Ahmar et al., 2020). Transcription activator-like effector nucleases (TALENs) is the highly precise, customizable, and flexible genome editing tool that enables highly specific and accurate editing of DNA (Bhardwaj and Nain, 2021). Zinc finger nucleases (ZFNs) are two component genome editing tools which are chimeric and synthetic with endonuclease systems and can insert or delete the gene of interest (Majumdar et al., 2022). Genome editing techniques result in transgenic breeding and precision breeding and thus revolutionizes conventional breeding in a great extent (Gao, 2021).
CRISPR/Cas9 is a cutting-edge genome-editing tool that enables the successful editing and modification of plant genomes, facilitating the introduction of new and improved traits to crop plants (Nekrasov et al., 2017). CRISPR/Cas9 is an RNA-guided DNA endonuclease, originating from the adaptive immune system of bacteria and archaea (Jakhanwal et al., 2021). The system consists of a single-guided RNA (sgRNA) composed of twenty nucleic acids and a Cas9 nuclease (Mehta and Merkel, 2020). CRISPR/Cas9 system targets the nucleic acids of invading pathogens or plasmids in bacteria and archaea (Ali et al., 2015). In this classical mechanism, the Cas9 endonuclease is directed by the sgRNA to cleave the target gene’s DNA strands’ protospacer adjacent motif (PAM) sequences (NGG sequences) (Zhang et al., 2021a). Genome editing of crop plants via CRISPR/Cas9 has been employed to create new crop varieties, improve existing traits of crops, investigate gene functions, improve yield production of crops, develop enhanced virus and pest-resistant crop varieties, and introduce salt and drought-resistant crop varieties (Nekrasov et al., 2017).
Keeping the alarming increase of population and future scarcity of food in mind, the aims of this review study are to discuss how CRISPR/Cas9 genome editing tool dramatically improved agricultural crops to date, expected future developments of crops utilizing this genome editing tool, and challenges to reach expected goals.
2. Evolution in crop advancement via CRISPR/Cas9
Genome editing technology has been focused on crop development. CRISPR/Cas9 has been employed to improve yield and quality, generation of abiotic and biotic stress resistance, and transgene-free genome-edited crops. Figure 1 illustrates the key revolutionary role of plant’s genome editing via CRISPR/Cas9 genome editing tool.
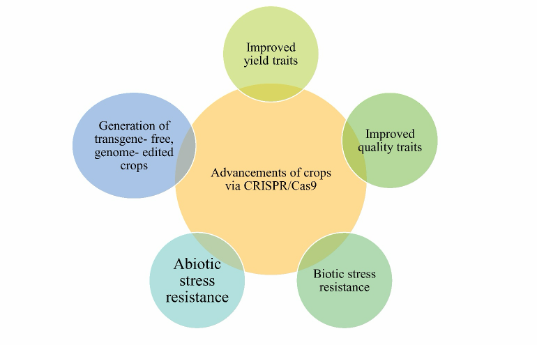
2.1 Improved yield traits
Yield is a complex and very important trait of agriculture. Oryza sativa L., commonly known as Asian cultivated rice is one of the most important food crops in the world, feeding over half of the global population. Consequently, high yield has become one of the major objectives of breeders and growers, with the development of hybrid rice improving the yield potential of the crops (Li et al., 2016; Zhou et al., 2012). Several new rice varieties with high-yield traits have been developed by editing three genes called OsPIN5b, OsMYB30, and GS3 using CRISPR/Cas9 (Zeng et al., 2020). In indica rice, knockout of the OsHXK1 gene by CRISPR/Cas9 tool results in improved yield and enhanced photosynthesis (Zheng et al., 2021). In maize, yield traits are associated with meristem size and controlled by CLE peptide signals. These traits can be improved in maize through CRISPR/Cas9-mediated promoter editing of CLE gene (CLE genes weak promoter alleles and a partially redundant compensating CLE gene which is newly identified) (Liu et al., 2021). Nitrogen is an essential nutrient for crops. In bread wheat (Triticum aestivum), the exact function of the ARE1 gene is still unknown. However, recent research has shown that wheat mutants generated through CRISPR/Cas9 editing exhibit increased nitrogen use efficiency under lower nitrogen conditions, leading to increased yields (Zhang et al., 2021b).
2.2 Improved quality traits
The quality trait is referred to as one of the most important targets for farmers and breeders. An increase in food quantity is important to meet global demand. However, quality of food is equally essential to ensure that it is supplied in greater amounts without potential health hazards and retains its full nutritional value (Zhang et al., 2018). A highly toxic heavy metal named cadmium has a serious health effect and affects human and animal health. A major source of dietary cadmium is rice which contains excessive amounts of cadmium from contaminated paddy fields. The CRISPR/Cas9 system effectively develops new rice lines with low cadmium accumulation without affecting rice yield. OsNramp5 is the metal transporter gene that has been knocked out to decrease cadmium content through the CRISPR/Cas9 system. CRISPR/Cas9 system induces OsNramp5 gene’s site-specific mutagenesis (Tang et al., 2017). Steroidal glycoalkaloids (SGAs) α-solanine and α- chaconine are accumulated in most potatoes. SGAs are highly toxic compounds and show toxicity against various organisms. In addition, it confers a bitter taste in humans. Mutations caused by the CRISPR/Cas9 system result in the silencing of several SGA biosynthesis genes and thus lead to a reduction in SGAs level. Here, knock out of the St16DOX gene which encodes a steroid 16α-hydroxylase in SGA biosynthesis causes the complete repeal of SGAs accumulation in hairy roots of potato (Nakayasu et al., 2018).
In tomato (Solanum lycopersicum), soluble sugar accumulation is inhibited by two genes called SlVPE5 and SlINVINH. Soluble sugar content is important for sweetness which is a major trait of tomatoes. So, editing of the above-mentioned genes is the main focus for improving soluble sugar content. Researchers successfully used the CRISPR/Cas9 tool to knock out these two genes and CRISPR-invinh1 or CRISPR-vpe5 (two genes’ knocked-out lines) was obtained via CRISPR/Cas9 (Wang et al., 2021a). Researchers found that in maize, both male fertility and pollen development genes have been successfully edited via the CRISPR/Cas9 genome-editing tool (Liu et al., 2022).
2.3 Biotic stress resistance
CRISPR/Cas9 genome editing system can also be utilized to offer molecular immunity against DNA viruses or RNA viruses in agricultural crop plants. For DNA viruses such as Tomato yellow leaf curl virus (TYLCV), Mungbean yellow mosaic virus (MeMV) and Beet curly top virus (BCTV), CRISPR/Cas9 system precisely targets multiple regions of viral genome. Single-guided RNAs (sgRNAs) target the coat protein (CP), intergenic region (IR), and replication associated protein (Rep) of viral genome and thus causes double stranded breaks in viral genome (Ali et al., 2015). Recent studies showed that for geminivirus replication, an essential element is the stem-loop intergenic sequence. For the creation of geminivirus-resistant crops, important targets are intergenic sequences (Ali et al., 2016).
In Arabidopsis thaliana to improve resistance to powdery mildew, EDR1 is an ideal target. Generally, EDR1 has three homologs. Cloning of the TaEDR1 gene from hexaploidy wheat showed higher levels of similarity among the three homologs of EDR1. CRISPR/Cas9 technology is used to generate Taedr1 plants by simultaneous modification of the three homologs of EDR1 genes. These Taedr1 wheat plants show increased resistance in Arabidopsis thaliana to powdery mildew and reduce yield loss successfully (Zhang et al., 2017).
Small brown planthopper (SBPH: Laodelphax striatellus Fallén) transmits rice black-streaked dwarf virus (RBSDV). It results in black-streaked dwarf disease in rice and causes potential loss in Chinese rice production (generally 10-40%). Recent studies showed that editing of eukaryotic translation initiation factor 4G (eIF4G) loci in Oryza sativa var. japonica cv. Nipponbare generates mutant rice varieties resistant to RBSDV (Wang et al., 2021b).
The two most devastating diseases of rice are bacterial blight and blast. Longke638S (LK638S) is a thermosensitive genic male sterile (TGMS) rice line. Bsr-d1, Pi21, and ERF922 are three important blast-resistance genes. CRISPR/Cas9-induced mutations of these genes generate Indica rice varieties with both bacterial blight and blast resistance traits (Zhou et al., 2021).
Cotton leaf curl disease (CLCuD) is caused by the begomoviruses. It is a member of the virus family Geminiviridae and has a devastating impact on cotton production. The CRISPR/Cas9 system is an extensive tool to control geminiviruses. In this system, Cas9 endonuclease is directed by sgRNA which can induce a precise target at a cleavage site (Iqbal et al., 2016). In the cucumber (Cucumis sativus L.), the eIF4E (eukaryotic translation initiation factor 4E) gene has been edited successfully via the CRISPR/Cas9 genome-editing tool. Editing of eIF4E loci generates cucumber varieties with broad virus resistance (Chandrasekaran et al., 2016). TYLCV (Tomato yellow leaf curl virus) is one of the most harmful viruses of tomatoes and reduces yield by up to 100%. Tomato is a globally important food with greater amount of trade value and have a role in controlling world hunger. Damage from this horticultural crop by virus is a devastating scenario. However, TYLCV virus is uncontrollable via conventional disease management approaches. Single-guided RNA (sg-RNAS) of CRISPR/Cas9 system targets coat protein (CP) and replication associated proteins (Rep) of the TYLCV virus genome, confers resistance against this virus and thus maintain the desired production of tomato for world population. Nicotiana tabacum and Tomato (Solanum lycopersicum) plants are recently engineered via CRISPR/Cas9 technology. The cap protein (CP) sequence of TYLC of tomato was efficiently targeted by CRISPR/Ca9 and it provided effective resistance to viruses in all homozygous tomato plants for T2 to T3 generations. However, CRISPR/Cas9 targets Rep genome less efficiently. So, these plants confer stable resistance against TYLCV for several generations (Tashkandi et al., 2018).
2.4 Experimental animal
Major abiotic stress is drought which challenges food security. Recent work shows that CRISPR/Cas9 can successfully edit the trehalase gene and thus decrease the activity of this gene and increase the accumulation of trehalose (α-D-glucopyranosyl-1, 1-α-D-glucopyranoside) in plants. Genome-edited crops generated by this process are more tolerant to drought stress (Nuñez-Muñoz et al., 2021).
In indica rice cultivar MTU1010, the drought and salt tolerance (DST) gene can be mutated via CRISPR/Cas9 genome editing technique to generate indica mega rice with enhanced salt and drought tolerance (Santosh Kumar et al., 2020). In maize (Zea mays), reduced ethylene sensitivity is proved to be shown by a gene called ARGOS8 and thus this gene acts as a negative regulator. In addition, improved yield production under drought resistance is also possible by the overexpression of the ARGOS8 gene. Examination shows that the mRNA expression level of the endogenous ARGOS8 gene is very low. CRISPR/Cas9 technology has proved to be the most useful tool to enhance the expression of the ARGOS8 gene in maize (Shi et al., 2016).
SlMPK3 is a mitogen-activated protein kinase or more precisely a serine-threonine kinase associated with stress tolerance in tomatoes. A conserved Thr- Glu- Tyr (TEY) motif is carried by SlMAPK3. Activation of the SlMAPK3 gene is performed by pathogen infections and ultraviolet-B radiation. In tomatoes, the SlMAPK3 gene’s function study is very important to understand its clear responses to drought conditions. The CRISPR/Cas9 technology is widely utilized to produce SlMAPK3 mutants in tomatoes (Wang et al., 2017).
UV-B is very harmful to plants and it is predominant. A UV-B photoreceptor regulating various aspects of plant morphogenesis is UVR8 (UV resistance photoreceptor 8) and its function is well-studied in Arabidopsis. In contrast, UVR8 in tomatoes is known as SIUVR8 and its function is still unknown. Recent research showed that the CRISPR/Cas9 genome editing technique successfully generated knock-out SIUVR8 mutant varieties of tomatoes. These edited varieties have been shown to control the accumulation of anthocyanin and hypothetical elongation in tomatoes under extreme UV-B stress conditions (Liu et al., 2020).
2.5 Generation of transgene-free, genome-edited crops
Half of the global population depends on Asian cultivated rice (Oryza sativa L.) for food. CRISPR/Cas9-mediated genome editing technology has been successfully applied to develop rice with improved yield and quality, without the introduction of transgenes (Tang et al., 2017; Zhou et al., 2016). Researchers successfully generated transgene-free rice varieties with carotene-enriched grains via CRISPR/Cas9 technology without off-target mutations and without hampering its morphology or yield (Dong et al., 2020). Optimizing the CRISPR/Cas9 genome editing technology is crucial and acts as a game-changer for crop improvement and precision breeding to establish sustainable agriculture and meet global food security. For the optimization of the CRISPR/Cas9 system, it is important to avoid transgene integration into the cells and reduce the target mutations. The CRISPR/Cas9 ribonucleoprotein (RNP) complexes are successfully used to generate transgene-free hexaploid bread wheat. Here 100 immature wheat embryos are generated from four to five independent mutations within seven to nine weeks (Liang et al., 2017). The Semidwarf phenotype of maize is associated with improved lodging resistance and improved product. The gibberellin oxidase gene contributes to this phenotype. Editing of this “green revolution” gene (Gibberellin-Oxidase20-3) with CRISPR/Cas9 has produced foreign – DNA-free semidwarf maize (Zhang et al., 2020). C to T base conversion is directed via CBEs (Cytidine base editors) which is a CRISPR/Cas9 derived tool. In tomatoes and potatoes, the acetolactate synthase (ALS) gene is targeted via CBE using transformation mediated by Agrobacterium. Researchers successfully edited cytidine bases and generated transgene-free chlorsulfuron-resistant potato and tomato via CRISPR/Cas9 cytidine base editor transformation by Agrobacterium (Veillet et al., 2019).
So, we can conclude that the CRISPR/Cas9 genome editing tool not only improves crops by genome editing but also generates transgene-free crops.
3. Challenges for the CRISPR/Cas9 genome editing technology
As we have discussed before, the CRISPR/Cas9 system has many potential applications in agricultural crop improvement. However, recent studies showed that CRISPR/Cas9 technology has some pitfalls or limitations.
Unexpected mutations are the biggest problems of the CRISPR/Cas9 system which is induced by off-target effects. Off-target cleavage may be caused by an inappropriate concentration ratio between sgRNA (single-guide RNA) and Cas9. The effect of off-target cleavage rises with the rise of improper concentration of Cas9: sgRNA ratio (Kleinstiver et al., 2016; Hsu et al., 2013). Undesired cleavage of DNA regions may also be caused by promiscuous PAM sites. Guide- RNA: Cas9 complexes when existing in higher concentration, cause the cleavage of off-target sites within or near the PAM (protospacer adjacent motif). But at low Cas9: guide- RNA concentration, cleavage of DNA may not occur (Pattanayak et al., 2013). For existing alignment tools, the Cas9/sgRNA target site is very short which is important for the identification of potential off-target sites effectively. To find off-target sites in any given genome, CasOT is a local tool (Xiao et al., 2014). The potential design of single guide RNA (sgRNA) is very important for genome editing via CRISPR/Cas9. CRISPR/Cas9 system activates the target gene in some cases, and in some cases, it inactivates a gene. The design of efficient sgRNA is assumed to be a major pitfall in this technology because it depends on various factors and varies within different sgRNAs (Moreno-Mateos et al., 2015; Xu et al., 2015; Doench et al., 2014). Recalcitrant sgRNA/target is another important challenge of the CRISPR/Cas9 system. Some factors, such as the target loci’s chromatin states and the sgRNA’s hairpin structures, may result in low efficiencies in certain sgRNAs (Shan et al., 2014).
Though the improvement of the efficiency of sgRNA and reducing the off-target effects of CRISPR/Cas9 is quite challenging, several strategies can be utilized to achieve these goals to some extent. Maintaining the GC content of sgRNA between 40-60% ensures the stability of the DNA, RNA complex and thus reduces the off-target activity and increases the on-target activity. sgRNAs with sequence length smaller than 20 nucleotides have greater specificity. Additionally, GG20 technique and chemical modification of sgRNA (incorporating 2′-O-methyl-3′-phosphonoacetate’ to the ribose-phosphate background of sgRNAs’ specific site) make sgRNAs more efficient. High fidelity Cas9 (HF-Cas9) is a modified version of Cas9 which reduces off-target cleavage of target DNA. Prime editors use prime editing guide RNA (PegRNA), Cas9 nickase (nCas9) and reverse transcriptase enzyme to effectively induce on-target genome editing in target genome. Optimized delivery of sgRNA and Cas9 into target cells also reduces the off-target effects. Another approach to reduce off-target effects of this genome editing tool is to reduce the concentration of Cas9: sgRNA (Mengstie et al., 2024; Ali et al., 2015).
4. Future expectancies of the CRISPR/Cas9 tool for improving important agriculturally important crop plants
CRISPR/Cas9 technology has been involved in various fields of agriculture. Further research can play an important role in the domestication of wild plants, the improvement of traits, and the improvement of delivery systems. These future expectancies of CRISPR/Cas9 for improving crops are summarized in Figure 2.

4.1 CRISPR/Cas9 technology for accelerating the domestication of wild plants
African rice (Oryza glaberrima and Oryza sativa landraces) has high pest and stress-resistance traits but lacks domestication traits such as lodging, seed shattering, and seed yield. Recently researchers successfully domesticated the African rice variety by disrupting the gene called HTD1 (which minimizes lodging). GS3, GWN1A, and GW2 are three genes controlling grain or yield traits, editing of these genes via CRISPR/Cas9 domesticate African wild rice (Lacchini et al., 2020). Tomato (Solanum lycopersicum; SI) has many wild relatives. Genome editing can also be used to domesticate tomatoes. Intensive breeding cycles are used by the breeders to cultivate modern tomato cultivars, but these cultivars are often by many biotic and abiotic stresses. In addition, it also results in loss of genetic diversity and fitness penalties. Wild tomato varieties are naturally stress-tolerant and can provide target genes for the domestication of wild tomatoes via accurately editing the genes that are responsible for tomato domestication (Li et al., 2018; Zsögön et al., 2018).
It is hoped that in the future, domestication of many other important wild and semi-domesticated crops will be possible. These new domesticated crops should have more stress tolerance and higher nutritional value with higher yields, and these should promote agricultural diversity and help to establish sustainable development in agriculture.
4.2 CRISPR/Cas9 system for future improvement of traits in agriculturally important crops
Utilization of CRISPR/Cas9 technology for future crop plant improvement, the root system is an important target. The root is important because of its role in stress tolerance and reproduction (Ahmadi et al., 2014). The CRISPR/Cas9 tool is a powerful tool for gene function analysis and crop improvement. Root editing is important in cereal crops for taking nutrients and water from the soil. CRISPR/Cas9 system offers precisely targeted mutagenesis which plays an important role in functional genomics studies in legume research. CRISPR/Cas9 technology will surely play a role in future crop improvement through the modulation of nodule and root genes linked to economically important legumes (Sun et al., 2015).
Genome editing tool also focuses on increased productivity and breeding of orphan crop species. Orphan crop species mainly have undesirable characteristics, and these crops resemble wild relatives. CRISPR/Cas9 system has also been used to mutate orthologues of tomato domestication. In addition, this genome editing tool also helps to improve genes that control plant architecture, fruit size, and flower production (Lemmon et al., 2018).
CRISPR/Cas9 system-mediated targeted genome editing will open many more amenities for the rapid and efficient modification of crops to boost yields, enhance nutrient content, increase stress tolerance, and protect against pests and diseases (Jones, 2015).
4.3. CRISPR/Cas9 technology for improved delivery systems
Recently the transformation of major monocots including maize has become possible. Transformation efficiency in maize and other monocots is mainly enhanced by the overexpression of the maize (Zea mays) Baby boom (Bbm) and maize Wuschel2 (Wus2) genes. Studies showed that this overexpression of these developmental genes (Bbm and Wus2) facilitates high transformation frequencies in a large number of maize inbred lines that were previously non-transformable (Xu et al., 2017; Lowe et al., 2016). In addition, morphogenic regulators Baby boom and Wuschel2 facilitate the transformation of sorghum varieties (Nelson-Vasilchik et al., 2018).
The favorable method for wheat transformation requires callus culture and regeneration and for this reason, it does not apply to many other cultivars. The Planta transformation method is very useful to avoid several problems that are associated with tissue culture and regeneration. Recent studies also discussed that some reliable targets for genetic transformation are egg cells, zygotes, sperm cells. The meristematic tissues of mature embryos can be successfully used in the planta wheat transformation system which utilizes a biolistic particle delivery system. Shoot apical meristems can also be used for delivery because these cells are ultimately differentiated into gametes (Hamada et al., 2017).
It is hoped that in the near future, CRISPR/Cas9 genome editing technology will facilitate more efficient delivery systems for genome editing.
5. Discussion
CRISPR/Cas9 (clustered regularly interspaced short palindromic repeats) is a versatile, cost-effective, simple, and highly accurate genome editing mechanism used widely in diverse fields including nanotechnology, agriculture, fisheries, environmental sciences, medicine (Ansori et al., 2023). This genome editing tool also has high- efficiency and high- specificity in site-directed mutagenesis or targeted mutagenesis. This technique enables the mutation of multiple genes or sometimes a single gene, including both homologous and non-homologous genes, through a transformation event. Mutant libraries are also constructed very efficiently using this technique. Transgene libraries are essential for the study of crop genomics as well as improvement of desirable traits of crops including biotic and abiotic stress resistant crops, improved yield and nutritionally rich crops (Zhang et al., 2019).
Researchers and breeders have already adopted these Cas9-based technologies in agriculturally important crop plants (Zheng et al., 2021). In addition, these tools provide a precise and efficient analysis of plant biology and enable plant science researchers to improve crop plants for food security. Researchers have already applied this technology to increase crop yield traits, improve quality traits, generate stress-resistant crop plants, speed hybrid breeding, generate transgene-free genome-edited crops (Nekrasov et al., 2017).
CRISPR/Cas9 system also faces some challenges, or this system has some pitfalls in crop genome editing. But the limitations are very few and researchers are trying to overcome these limitations (Chen et al., 2019). Genome editing using CRISPR/Cas9 is prone to off-target mutations which can result in undesirable edits in non-target genes and develop harmful traits and products in crops (Han et al., 2020). Resistant genes integrated into crops for better outcomes may spread to wild relatives and result in loss of biodiversity. Additionally, plants with resistant traits such as pesticide and herbicide resistant plants can pose potential harm to non-target organisms or beneficial pests and results in super weeds respectively. There are ethical concerns about the consumption of GM foods. People question the ethics of developing genetically modified or edited foods and their impact on human health is still questionable (Pankaj and Kumar, 2023). Ethical regulations should be developed addressing the accuracy of the genome editing and safety concerns of the genome edited foods. Biodiversity preservation should also be a top priority for scientists and Biotech companies. Above all, raising public awareness about genetically modified foods and changing public perception about it is another way to ensure its sustainable use in agriculture (Katam et al., 2022; Gupta et al., 2021). It is also hoped that this genome editing technology will play more precise and important roles in crop genome editing to improve crops in the near future. Undoubtedly, the CRISPR/Cas9 genome editing tool is one of the most important and powerful editing tools for the overall improvement of crops and thus it supplies food for the increasing population and reduces world hunger.
6. Conclusion
Food security is the biggest challenge for the rapidly growing global population. The increase in food production using the available limited resources requires alternative methods to conventional agriculture and breeding. Climate change and harsh environment make it a huge challenge. Genome editing technologies have emerged as solutions for this crisis and among various genome editing technologies CRISPR/Cas9 is a fast-growing tool in the genome editing sector that is versatile and makes perfect edits in the target genome. Various food crops such as tomato, rice, wheat, maize, potato has already been modified using CRISPR/Cas9, and these modified varieties are superior versions of their original varieties with desired traits. However, genome editing technologies like CRISPR/Cas9 raises concerns for ethical issues, biosafety regulations, and ecological safety. People are still not ready to have genetically modified (GM) foods due to health concerns and perceptions. It may negatively impact the adoption of CRISPR/Cas9 in food crop improvement. Furthermore, ownership of genome edited crops raises another question about equal cultivation of these crops or it’s monopolization by big Bioytech companies. Development of resistant crops may result in gene flow and crossbred with wild plants and thus imbalance biodiversity. Following biosafety guidelines is another concern that could hinder the adoption of CRISPR/Cas9 in future. However, by establishing and following appropriate biosafety regulations, raising awareness about genome edited crops, clarifying the ethical issues regarding it, and mitigating ecological risk can help to get global approval and adoption of CRISPR/Cas9. It is hoped and believed that in the future, this genome editing tool will revolutionize our food sector and pave the path for sustainable agriculture.
Acknowledgements
The Author acknowledge the contribution of Islamic University, Kushtia, Bangladesh for their funding and assistance all member of Laboratory of Medical and Environmental Biotechnology, Department of Biotechnology and Genetic Engineering, Islamic University, Kushtia, Bangladesh.
Funding information
Special Allocation by Islamic University funding ID: Ref.- 141/Edu./IU-27 Date: 5-11-2024, Financial Year: 2023-24.
Ethical approval
As this study is a literature review, no ethical approval is needed.
Data availability
Not applicable.
Informed consent statement
Not applicable.
Conflict of interest
The authors declare no competing interests.
Authors’ contribution
Conceptualization and lead the research: Masuma Anzuman; Formatting and design the work: Mohammad Abu Hena Mostofa Jamal; Written the application part: Md. Khasrul Alam; Written the background and discussion of the study: Md. Rezuanul Islam; Supervision: Nilufa Akhter Banu.All authors critically reviewed the manuscript and agreed to submit final version of the article.


