Newcastle disease (ND) is one of the most highly contagious and lethal viral diseases affecting poultry, posing a significant global concern (Suarez et al., 2020 ![]() ; Vanmechelen et al., 2022
; Vanmechelen et al., 2022 ![]() ). Approximately 250 avian species across 27 of the 50 bird groups are known to be susceptible to natural or experimental infection with Newcastle disease virus (NDV). It is likely that many additional species are vulnerable but have not yet been identified (Suarez et al., 2020
). Approximately 250 avian species across 27 of the 50 bird groups are known to be susceptible to natural or experimental infection with Newcastle disease virus (NDV). It is likely that many additional species are vulnerable but have not yet been identified (Suarez et al., 2020 ![]() ; Smietanka et al., 2014
; Smietanka et al., 2014 ![]() ). ). The World Organization for Animal Health (OIE) designates Newcastle disease virus (NDV) infection as a notifiable disease. The disease was first reported in poultry in 1926 in Java, Indonesia, and Newcastle upon Tyne, England, and has since become widespread globally. NDV, formally known as avian paramyxovirus sero type-1 (APMV-1), belongs to the genus Avulavirus, within the family Paramyxoviridae and the order Mononegavirales (Li et al., 2019
). ). The World Organization for Animal Health (OIE) designates Newcastle disease virus (NDV) infection as a notifiable disease. The disease was first reported in poultry in 1926 in Java, Indonesia, and Newcastle upon Tyne, England, and has since become widespread globally. NDV, formally known as avian paramyxovirus sero type-1 (APMV-1), belongs to the genus Avulavirus, within the family Paramyxoviridae and the order Mononegavirales (Li et al., 2019 ![]() ). NDV is an enveloped, linear, non-segmented, negative-sense single-stranded RNA virus. Its genome comprises six genes and spans 15,186 nucleotides (Maes et al., 2019
). NDV is an enveloped, linear, non-segmented, negative-sense single-stranded RNA virus. Its genome comprises six genes and spans 15,186 nucleotides (Maes et al., 2019 ![]() ). These genes encode nucleoprotein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN) protein, and large protein (L), organized in the sequence 3′-NP-P-M-F-HN-L-5′.
). These genes encode nucleoprotein (NP), phosphoprotein (P), matrix protein (M), fusion protein (F), hemagglutinin-neuraminidase (HN) protein, and large protein (L), organized in the sequence 3′-NP-P-M-F-HN-L-5′.
The pathogenicity of NDV strains can vary significantly based on the host species. Among poultry, chickens and turkeys are the most susceptible to NDV, while ducks and geese are the least susceptible and are often considered carriers that are resistant to even the most lethal strains for chickens (Rehan et al., 2019 ![]() ).
).
The occurrence of NDV outbreaks in vaccinated flocks indicates the emergence of more virulent strains, leading to high morbidity and mortality rates among the birds and causing significant economic losses. This underscores the importance of regularly assessing circulating NDV strains to develop effective management measures to prevent such infections. NDV strains are categorized into five pathotypes: viscerotropic velogenic, neurotropic velogenic, mesogenic, lentogenic, and asymptomatic. Depending on the specific pathotype, as well as the host's physiology and immune status, the virus can damage the gastrointestinal, pulmonary, and neurological systems (Wiseman et al., 2018 ![]() ).
).
Pathotypically, NDV strains can be classified into three groups based on their pathogenicity: lentogenic, mesogenic, and velogenic (Dimitrov et al., 2016 ![]() ). Velogenic and mesogenic strains are considered virulent NDVs, while lentogenic strains are characterized by low virulence. Pathotyping of NDV strains involves assessing their virulence using methods such as the intracerebral pathogenicity index (ICPI) in 1-day-old chicks, the intravenous pathogenicity index (IVPI) in 6-week-old hens, and the mean death time (MDT) in embryonated chicken eggs (ECE) (Mphuthi et al., 2018
). Velogenic and mesogenic strains are considered virulent NDVs, while lentogenic strains are characterized by low virulence. Pathotyping of NDV strains involves assessing their virulence using methods such as the intracerebral pathogenicity index (ICPI) in 1-day-old chicks, the intravenous pathogenicity index (IVPI) in 6-week-old hens, and the mean death time (MDT) in embryonated chicken eggs (ECE) (Mphuthi et al., 2018 ![]() ; Hossain et al., 2017
; Hossain et al., 2017 ![]() ).
).
Genetically, NDVs are divided into two classes: class I and class II. Class II strains have been isolated from both wild and domestic birds, encompassing both virulent and non-virulent variants, and include at least 18 genotypes (Absalón et al., 2019 ![]() ). Class I NDV strains are primarily found in wild birds and typically have low virulence. Previously, these strains were categorized into at least nine genotypes. In the current classification system, they are grouped into a single genotype with at least three sub-genotypes (Diel et al., 2012
). Class I NDV strains are primarily found in wild birds and typically have low virulence. Previously, these strains were categorized into at least nine genotypes. In the current classification system, they are grouped into a single genotype with at least three sub-genotypes (Diel et al., 2012 ![]() ). The molecular pathotyping of various strains, often achieved through F gene analysis and reverse transcription PCR (RT-PCR), has been instrumental in comprehending and categorizing NDV strains based on their virulence into the aforementioned three categories (Liu et al., 2016
). The molecular pathotyping of various strains, often achieved through F gene analysis and reverse transcription PCR (RT-PCR), has been instrumental in comprehending and categorizing NDV strains based on their virulence into the aforementioned three categories (Liu et al., 2016 ![]() ). Commonly employed methods for detecting and typing NDV strains include virus isolation, immunohistochemistry, RT-PCR, gene sequencing, and microarrays (Souf, 2016
). Commonly employed methods for detecting and typing NDV strains include virus isolation, immunohistochemistry, RT-PCR, gene sequencing, and microarrays (Souf, 2016 ![]() ).
).
The primary preventive measures for Newcastle disease (ND) include maintaining hygiene and implementing vaccination protocols. Humans, like avian species, can also be affected by ND (Ali et al., 2021 ![]() ; Ali et al., 2019
; Ali et al., 2019 ![]() ). Exposure to high levels of Newcastle disease virus (NDV) can lead to conjunctivitis in humans. Among infectious diseases, ND is particularly notorious for causing significant economic losses in poultry and related products (Ali et al., 2021
). Exposure to high levels of Newcastle disease virus (NDV) can lead to conjunctivitis in humans. Among infectious diseases, ND is particularly notorious for causing significant economic losses in poultry and related products (Ali et al., 2021 ![]() ; Yusoff and Tan, 2001
; Yusoff and Tan, 2001 ![]() ).
).
Newcastle disease virus (NDV) can be confirmed using several diagnostic tests including hemagglutination (HA) and hemagglutination inhibition (HI) tests, virus neutralization tests, enzyme-linked immunosorbent assays (ELISA), plaque neutralization tests, and reverse-transcriptase polymerase chain reaction (RT-PCR) (Ali et al., 2021 ![]() ; Wajid et al., 2017
; Wajid et al., 2017 ![]() ). Currently, the most commonly used method to assess the virulence of an NDV isolate involves RT-PCR combined with sequencing of the F cleavage site. Unlike traditional laboratory diagnostic methods such as viral isolation, RT-PCR is more sensitive, specific, and faster. Many laboratories utilize real-time RT-PCR (rRT-PCR), initially employing a primer and probe set to detect NDV, followed by a second primer and probe set to determine the virulence of the virus. This molecular approach forms the basis for evaluating NDV virulence in various laboratories (Ali et al., 2021
). Currently, the most commonly used method to assess the virulence of an NDV isolate involves RT-PCR combined with sequencing of the F cleavage site. Unlike traditional laboratory diagnostic methods such as viral isolation, RT-PCR is more sensitive, specific, and faster. Many laboratories utilize real-time RT-PCR (rRT-PCR), initially employing a primer and probe set to detect NDV, followed by a second primer and probe set to determine the virulence of the virus. This molecular approach forms the basis for evaluating NDV virulence in various laboratories (Ali et al., 2021 ![]() ).
).
Despite widespread use of vaccination programs in Bangladesh to prevent and control Newcastle disease (ND), the disease continues to be endemic throughout the country, posing a persistent threat to commercial poultry. Severe outbreaks of ND occasionally occur despite routine immunization of chickens with live NDV vaccines derived from mesogenic and lentogenic strains (Rahman et al., 2018 ![]() ). Differences in biology, serology, and genetics between the current strains of NDV and vaccine viruses are considered significant factors contributing to the frequent recurrence of the disease in vaccinated poultry flocks in Bangladesh. There has been limited research on the molecular and clinical characteristics of NDVs circulating in Bangladesh (Khokon et al., 2017
). Differences in biology, serology, and genetics between the current strains of NDV and vaccine viruses are considered significant factors contributing to the frequent recurrence of the disease in vaccinated poultry flocks in Bangladesh. There has been limited research on the molecular and clinical characteristics of NDVs circulating in Bangladesh (Khokon et al., 2017 ![]() ).
).
This study was conducted to detect and characterize Newcastle Disease Virus (NDV) isolated from the brain, trachea, and proventriculus of birds suspected to have died from ND, through post-mortem examinations conducted in Trishal and Mymensingh Sadars, Bangladesh. The primary objectives of this study include isolating and identifying NDV from infected layer chickens, confirming the isolated virus using Hemagglutination (HA) test and Reverse Transcription Polymerase Chain Reaction (RT-PCR), and characterizing the pathotype of the NDV field isolate using Mean Death Time (MDT) and Intracerebral Pathogenicity Index (ICPI) tests
2. Materials and Methods
2.1 Ethical approval
No ethical approval is required for this study.
2.2 Study area
The current study was conducted over a period of 20 months, spanning from July 2021 to March 2023, at the Laboratory of the Department of Microbiology and Hygiene, Faculty of Veterinary Science, Bangladesh Agricultural University (BAU), Mymensingh (Figure 1). The study was conducted in three selected upazilas (Jhenaidahsadar, Harinakunda, Kotchadpur) in Jhenaidah district of Bangladesh (Figure 1).

2.3 Experimental design
The entire study was divided into three steps. For this purpose, 12 samples consisting of brain, trachea, and proventriculus were collected from different farms at Trishal and Sadar of Mymensingh, showing positive signs and symptoms of ND virus. Following accurate labeling and aseptic preservation in a refrigerated box, the collecting sample was promptly delivered to the Department of Microbiology and Hygiene laboratory at BAU, Mymensingh, and stored at -20°C until tested. The virus was isolated by propagation into embryonated chicken eggs. A slide hemagglutination test identified the ND virus primarily. Finally, RT-PCR was used to validate the virus identification, and pathogenicity indices were used to determine the pathotype.
2.4 Fertile eggs of chicken
Brown and white shelled NDV seronegative chicken eggs were purchased from a healthy flock with a good hatching rate obtained from the local rustic area in Mymensingh. The eggs were disinfected with 70% ethanol then cleaned with tissue. The eggs were incubated at 37°C for 11 days, and turned manually twice daily. The Newcastle disease virus (NDV) was propagated using well-developed, viable embryos.
2.5 Molecular analysis
The Viral RNA extraction was conducted following the procedure of Liu et al. (2012 ![]() ) using Viral RNA extraction kit (Promega, USA). For reverse transcription, we used the Viral GoScript™ Reverse Transcriptase kit (Promega, USA) to efficiently synthesize first-strand cDNA from RNA templates (Anisimova et al., 2024
) using Viral RNA extraction kit (Promega, USA). For reverse transcription, we used the Viral GoScript™ Reverse Transcriptase kit (Promega, USA) to efficiently synthesize first-strand cDNA from RNA templates (Anisimova et al., 2024 ![]() ). Appropriate primer sequences were selected for PCR to amplify the genes of the ND virus. These primers targeted the gene of NDV and produced 970bp and 839bp respectively fragment of Fusion-Matrix gene and Fusion gene (Liu et al., 2003
). Appropriate primer sequences were selected for PCR to amplify the genes of the ND virus. These primers targeted the gene of NDV and produced 970bp and 839bp respectively fragment of Fusion-Matrix gene and Fusion gene (Liu et al., 2003 ![]() ) (Table 1).
) (Table 1).
Table 1. The sequences of the primers used for the PCR of the gene of NDV.
| Name of the Primer | Primer Sequence | Amplicon size (bp) | Reference | |
| (F-M) gene | F | 5’-GTGAAGCTTGAGTCTGTGAGTCGTAC-3’ | 970 (bp) | Liu et al.(2003) |
| R | 3’-GCCGAATTCCCGAATCATCACGACGCTTAA-5’ | |||
| Fusion gene (F) | F | 5’-TTAAGCTTGTAGTGGCTCTCATCTGATC-3’ | 839 (bp) | |
| R | 3’-CACCGGTACCCCTATTCTGATCG-5’ | |||
For virus isolation, 12 samples were collected from chickens exhibiting symptoms suggestive of Newcastle Disease (ND) virus during the study period. These samples were obtained from the brain, trachea, and proventriculus of clinically suspected deceased chickens through post-mortem examinations conducted in Trishal and Mymensingh Sadar, Bangladesh (Table 2).
Table 2. Number of collecting samples from different tissues of chickens.
| Location and no. of birds | Types of tissue | No. of samples | Total samples |
| Trishal (2 birds) | Brain | 2 | 12 samples of 04 NDV suspected chickens |
| Trachea | 2 | ||
| Proventriculus | 2 | ||
| Brain | 2 | ||
| Mym. Sadar (2 birds) | Trachea | 2 | |
| Proventriculus | 2 |
2.6 Measuring the cDNA concentration by NanoDrop spectrophotometer
The NanoDrop spectrophotometer assesses DNA, RNA, and protein concentrations using only a 2-µL sample on its pedestal. This enables rapid measurement of multiple samples in under a minute, contrasting with conventional spectrophotometers that require a 1-cm cuvette. DNA purity is evaluated by the ratio of absorbance at 260 nm and 280 nm, where a ratio around 1.8 indicates high purity. Ratios below 1.6 may indicate the presence of proteins, phenol, or other contaminants that absorb strongly at 280 nm.
2.7 Protocol used for PCR
The PCR master mixture, primer, Extracted DNA and Nuclease free water was put into the PCR cooler box. Required numbers of PCR tubes with labeling were also put into PCR cooler box. Then the reaction mixture was prepared by adding Nuclease free water, PCR master mixture, forward, and reverse primers in an Eppendorf tube (Table 2). Next, the reaction mixture was dispensed into each PCR tube. Subsequently, 5 µl of isolated DNA template was added to each tube and thoroughly mixed using pipette tips. Finally, the PCR tubes were placed into the thermocycler for amplification.
2.7.1 Electrophoresis of the PCR products
PCR product 5 µl, which contained loading dye, was loaded to the appropriate well of the gel with a micropipette. DNA ladder was loaded in one well. The electrophoresis was connected to the power supply and was run at 100V for 20 min, and then the supply switched off. The gel was placed on the ethidium bromide for 15 min and then put into water for 5 min. The image documentation system's dark chamber is then illuminated by a UV transilluminator. Finally, the image was seen, focused, and stored into a pen drive.
2.8 Statistical analysis and pathotypical characterization of isolated NDV
2.8.1 Determination of mean death time (MDT) in chicken embryo
MDT was determined as per the method of El-Morshidy et al. (2021 ![]() ). Mean Death Time (MDT) is calculated as the number of hours at which 100% mortality of embryos occurs in the highest dilution divided by the number of dead embryos observed. The equation is as follow,
). Mean Death Time (MDT) is calculated as the number of hours at which 100% mortality of embryos occurs in the highest dilution divided by the number of dead embryos observed. The equation is as follow,

2.8.2 Determination of intracerebral pathogenicity index (ICPI)
ICPI was ascertained according to El-Morshidy et al. (2021 ![]() ). The ICPI was derived using the mean score per bird observation over an 8-day period. The equation is as follow,
). The ICPI was derived using the mean score per bird observation over an 8-day period. The equation is as follow,

3. Results
3.1 Isolation and identification of Newcastle disease virus (NDV)
3.1.1 Clinical findings observed in NDV-infected chickens
Recorded clinical signs were watery greenish diarrheic feces, loss of appetite, respiratory distress (gasping and coughing), nasal discharge, dropping of legs and wings, tremors, high fever, severe depression and prostration followed by death which was recorded through interview with farm owners (Figure 2).

3.1.2 Gross lesions observed during post mortem examination
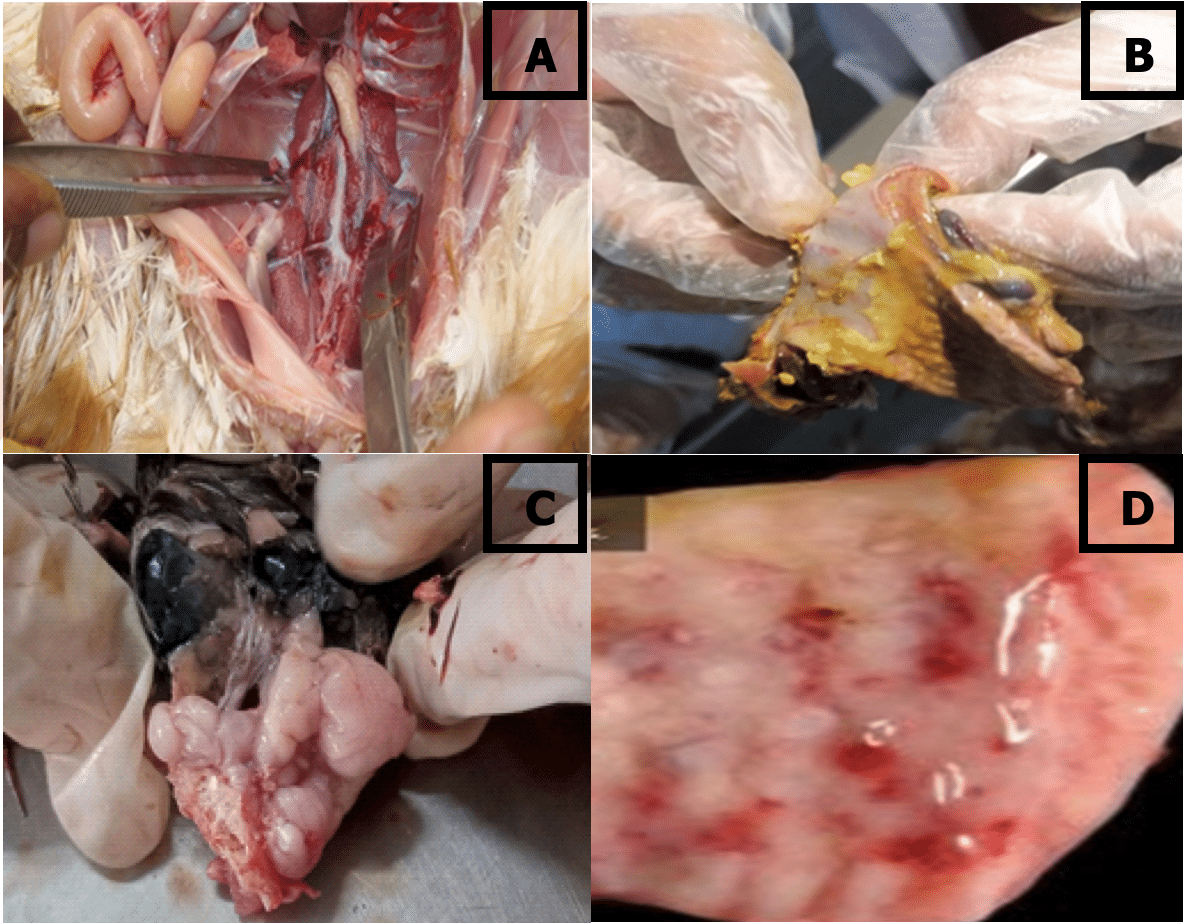
Hemorrhage in the proventriculus, button like ulcer in the duodenum, brush paint hemorrhage in the colon and congestion and hemorrhage in the trachea (Figure 3).
3.2 Isolation of NDV in chicken embryos
A total of 12 (brain, trachea, and proventiculus) samples were obtained and implanted into chicken embryos for virus isolation, where NDV generated hemorrhagic lesion, encephalitis, and embryo mortality. Allantoic fluid (AF) from embryos that died within 48 hours post-inoculation was collected. The presence of hemagglutinating virus was confirmed using a direct plate hemagglutination (HA) test with 2% chicken red blood cells (cRBC) on an HA plate. Among the 12 post-mortem samples obtained from naturally infected layer chickens, Newcastle Disease (ND) virus was detected in 4 samples using the HA test.
3.2.1 Inoculum preparation from tissue samples
The collected brain, trachea, and proventriculus were chopped into minute pieces with sterile scissors and forceps, and then ground with a sterile pestle and mortar with sea sand to make suspensions with sterile PBS. Tissue collected from chicken farms were put in a Petridish plate, washed in PBS, and then taken into an eppendrof tube. Then samples were centrifuged at 2500-3000 RPM for 15 min maintaining 4°C, and the supernatant was collected and then passed through in syringe filter and treated with an antibiotic (Streptopen) and stored at –20°C and -80°C for further use.
3.2.2 Inoculation of preparaing inoculum into embryonated eggs for virus propagation and allantoic fluid collection
Ten-day-old embryonated eggs were selected by candling to ensure viability. A point of inoculation was made on the side of each egg at the embryo site identified during candling. The top of the air sac and the marked point of inoculation were disinfected using iodine tincture. An electric dentist's drill machine was used to create a small hole near the top of the air sac. Approximately 0.1 ml of inoculum was then introduced through the allantoic cavity route. The eggs were then incubated at 37°C for 5 days and monitored twice daily. Any embryos that died within 24 hours of inoculation were excluded from the study. Only live embryos were kept for up to 5 days post-inoculation, and allantoic fluid was collected from these embryos.
3.2.3 Observation of embryo lesions infected with NDV field isolates
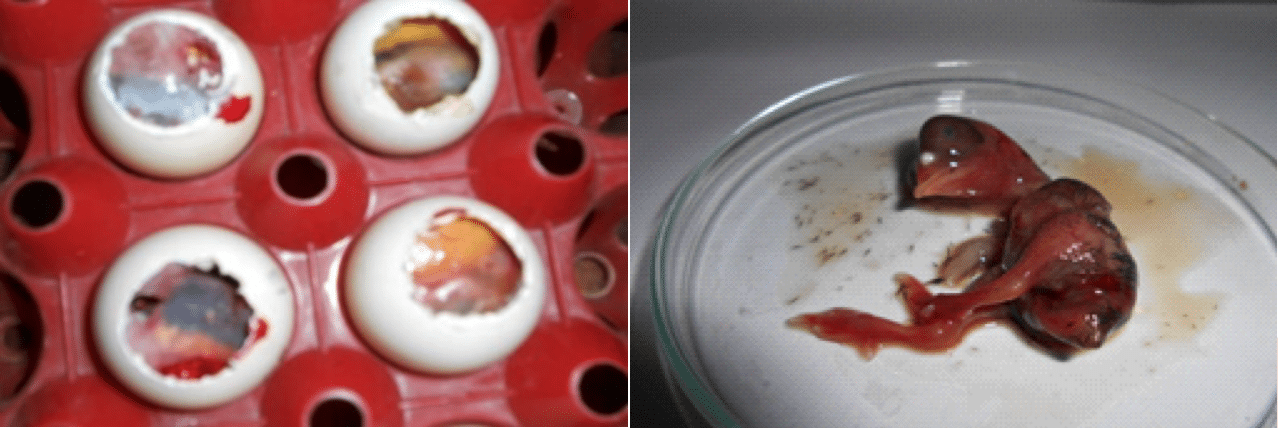
After virus inoculation into the embryonated chicken eggs, the allantoic fluid (AF) collected from the embryos those were found dead within 48 hours. Due to virus infection the embryonated chicken eggs were died and the dead embryos were hemorrhagic and edematous (Figure 4 and 5).

3.3 Confirmation the isolated NDV virus
For the propagation of the working virus, the collected samples were inoculated into chicken embryos and confirmed as ND virus by various tests and techniques. The virus was detected utilizing the slide HA approach using 2% chicken RBCs. All the isolate were HA positive (Figure 6).

3.3.1 Reverse transcription polymerase chain reaction (RT-PCR)
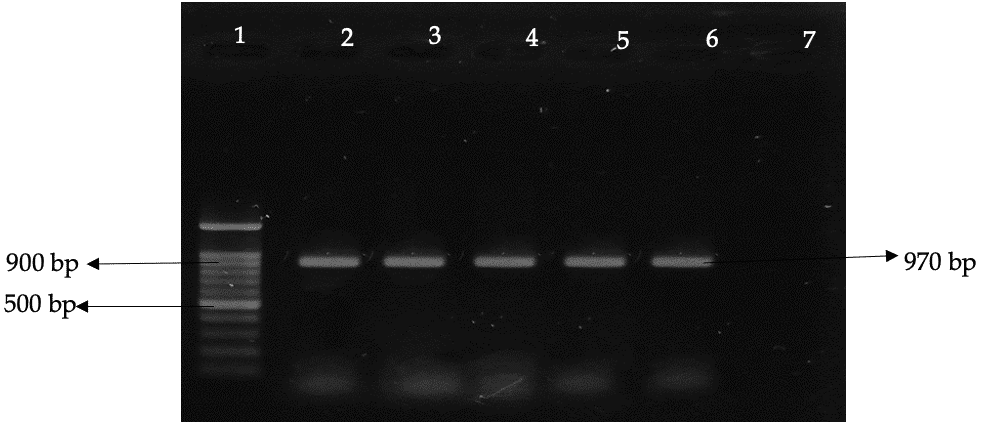
Oligo nucleotide primers were utilized to amplify 839 bp amplicons of the 'F' gene and 970 bp amplicons of the 'M' gene of NDV, which demonstrated equivalent sensitivity and specificity with RNA recovered from the allantoic fluid as with conventional NDV genomic RNA.
3.3.1.1 The measuring cDNA concentration by NanoDrop spectrophotometer
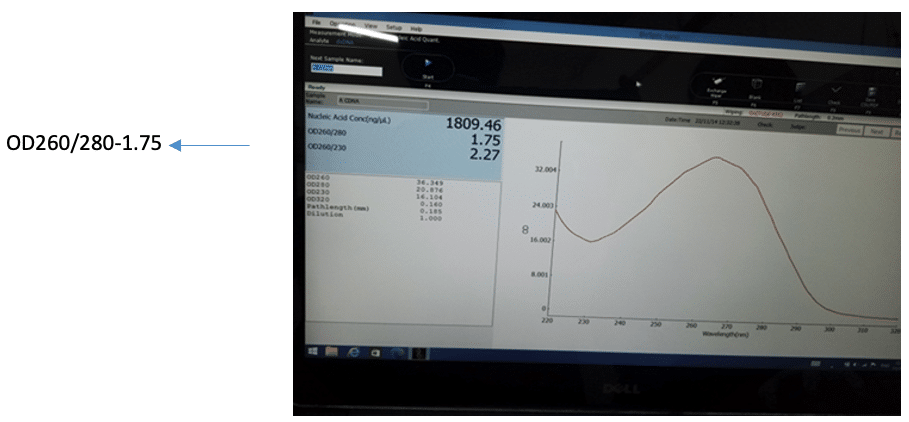
The ratio of absorbance at 260 nm and 280 nm is commonly used to assess DNA purity. Typically, DNA is considered "pure" when this ratio is around 1.8. A lower ratio, such as 1.6, may indicate the presence of proteins, phenol, or other substances that absorb strongly near 280 nm. In this case, an absorbance ratio of 1.75 suggests the purity of the cDNA (Figure 7).
3.3.1.2 ‘F’ gene detection by RT-PCR
The cDNA produced from the reverse transcription (RT) reaction was utilized to amplify a specific, larger-sized fragment of the 'F' gene through PCR, resulting in an amplicon of 839 base pairs. Fusion gene is known as the main determinant of pathogenicity. All the isolates were positive for ‘F’ gene (Figure 8).

3.3.1.3 ‘F’ and ‘M’ gene detection by RT-PCR
The isolates’ cDNA from the RT reaction was once more utilized to amplify the 'F' and 'M' protein genes (970 bp).Virus assembly uses the matrix gene. Every isolate displayed a successful detection of the ‘F’ and ‘M’ gene (Figure 9).
3.4 Pathotypical characterization of isolated NDV strains
3.4.1 Determination of MDT and ICPI of NDV field isolates
In vivo testing for virus pathogenicity includes determining the Mean Death Time (MDT) in chicken embryonated eggs and calculating the Intracerebral Pathogenicity Index (ICPI) using one-day-old chicks. Pathogenicity studies indicated that the Mean Death Time (MDT) was 54.34 hours, and the Intracerebral Pathogenicity Index (ICPI) value was 1.55 (Table 3 and 4).
On the basis of pathogenicity indices (MDT and ICPI), one of the virus isolate (NDV-1) found as velogenic which was found in field level of from Mymensingh district (Table 5).
Table 3. Calculation of MDT of NDV field isolate-1.
Table 4. Calculation of ICPI of NDV field isolate-1.
Table 5. Result of MDT and ICPI tests on NDV isolate-1
4. Discussion
Newcastle Disease (ND) poses a significant threat to the health and welfare of poultry globally. The OIE has classified it as a listed disease, mandating that outbreaks of mesogenic or velogenic ND be reported Anjum et al. (2020 ![]() ). Despite extensive vaccination efforts, ND remains endemic in numerous regions worldwide, including industrialized countries. Occasionally, outbreaks of NDV occur even in countries that have been declared free of the disease (Mayers et al., 2017
). Despite extensive vaccination efforts, ND remains endemic in numerous regions worldwide, including industrialized countries. Occasionally, outbreaks of NDV occur even in countries that have been declared free of the disease (Mayers et al., 2017 ![]() ).
).
Newcastle disease is a disease of domesticated and wild birds that causes considerable economic loss and hinders the development of the poultry industry. This disease is an acute, with high mortality in clinical cases. Infected birds are treated with a range of therapeutic treatments, but there is no suitable drug to prevent this viral disease. As a result, vaccination was the only effective method of disease control.
4.1 Isolation and identification of NDV
In the current study, samples were obtained from chickens exhibiting two different syndromes: one characterized by neurological signs including neck stretching, forward posture with legs backward, leg and wing drooping, leg paralysis, tremors, high fever, severe depression, and prostration leading to death; the other characterized by respiratory distress such as gasping and coughing, along with symptoms of nasal discharge, watery greenish diarrheic feces, and loss of appetite. These syndromes were also found by other researchers (Ragab et al., 2022 ![]() ; Deka et al., 2022
; Deka et al., 2022 ![]() ; ; Shanmuganathan et al., 2017
; ; Shanmuganathan et al., 2017 ![]() ). A total of 12 post-mortem samples (4 brains, 4 tracheae and 4 proventriculus) were collected for the collection of ND virus in avian embroys from Trishal and Mymensingh sadar in Bangladesh. Proventiculus samples were also found positive for NDV in the present study which also found by some workers (Balachandran et al., 2014
). A total of 12 post-mortem samples (4 brains, 4 tracheae and 4 proventriculus) were collected for the collection of ND virus in avian embroys from Trishal and Mymensingh sadar in Bangladesh. Proventiculus samples were also found positive for NDV in the present study which also found by some workers (Balachandran et al., 2014 ![]() ; Putri et al., 2022
; Putri et al., 2022 ![]() ). All 04 isolates were confirmed as ND virus by HA test and RT-PCR technique which related to the findings of Syamsiah Aini et al. (2022
). All 04 isolates were confirmed as ND virus by HA test and RT-PCR technique which related to the findings of Syamsiah Aini et al. (2022 ![]() ); Ahmadi et al. (2021
); Ahmadi et al. (2021 ![]() );and Orabi et al. (2017
);and Orabi et al. (2017 ![]() ). In Bangladesh, Hossain et al. (2017
). In Bangladesh, Hossain et al. (2017 ![]() ) assessed eleven NDV isolates derived from 19 field samples. Initially, all hemagglutination (HA)-positive allantoic fluid (AF) samples were collected and preliminarily confirmed using hemagglutination inhibition (HI) tests with anti-Avian Paramyxovirus-1 (APMV-1) polyclonal serum. Subsequently, RT-PCR with F gene-specific primers specific to NDV was employed for definitive virus confirmation, mirroring the methodology employed in the current study. The samples were collected in the same district but in different localities. MDT, ICPI, and IVPI indices indicated that all NDV isolates from 2011 and 2012 were velogenic, consistent with findings from the current investigation. Hossain et al. (2017
) assessed eleven NDV isolates derived from 19 field samples. Initially, all hemagglutination (HA)-positive allantoic fluid (AF) samples were collected and preliminarily confirmed using hemagglutination inhibition (HI) tests with anti-Avian Paramyxovirus-1 (APMV-1) polyclonal serum. Subsequently, RT-PCR with F gene-specific primers specific to NDV was employed for definitive virus confirmation, mirroring the methodology employed in the current study. The samples were collected in the same district but in different localities. MDT, ICPI, and IVPI indices indicated that all NDV isolates from 2011 and 2012 were velogenic, consistent with findings from the current investigation. Hossain et al. (2017 ![]() ) studied the prevalence of NDV across various poultry farms in nine localities spanning five districts, reporting higher rates of positive isolates and more severe pathogenicity outcomes compared to the present study. In this study, we collected samples only from the Mymensingh district. This variation could be related with the regional variation of samples.
) studied the prevalence of NDV across various poultry farms in nine localities spanning five districts, reporting higher rates of positive isolates and more severe pathogenicity outcomes compared to the present study. In this study, we collected samples only from the Mymensingh district. This variation could be related with the regional variation of samples.
4.2 Molecular characterization of NDV
RT-PCR is a speedy, strong, and highly specific molecular technology for confirmatory detection of several viruses, including Newcastle disease virus (Anjum et al., 2020 ![]() ). The development of molecular technologies has paved the door for quick and specific identification of infectious organisms, hence RT-PCR was used for identification of the ND virus. During the present study, isolates of NDV exhibit specific amplified product of 839 bp and 970 bp size respectively using primers Fusion-Forward/Reverse and Matrix- Forward/Reverse. These findings were confirmed by some researches (Miller et al., 2010
). The development of molecular technologies has paved the door for quick and specific identification of infectious organisms, hence RT-PCR was used for identification of the ND virus. During the present study, isolates of NDV exhibit specific amplified product of 839 bp and 970 bp size respectively using primers Fusion-Forward/Reverse and Matrix- Forward/Reverse. These findings were confirmed by some researches (Miller et al., 2010 ![]() ; Wise et al., 2004
; Wise et al., 2004 ![]() ). Other researchers have verified the legitimacy of the PCR product based on the size of the amplicons (Fazel and Mehrabanpour, 2018
). Other researchers have verified the legitimacy of the PCR product based on the size of the amplicons (Fazel and Mehrabanpour, 2018 ![]() ; Singh et al., 2005
; Singh et al., 2005 ![]() ). In the present study for ‘F’ and ‘M” gene of NDV detection, primer designed by Liu et al. (2003
). In the present study for ‘F’ and ‘M” gene of NDV detection, primer designed by Liu et al. (2003 ![]() ) was used in this study and the amplicon size respectively was 839 bp and 970 bp. The RT-PCR technique has been successfully used by numerous researchers (Sajo et al., 2022
) was used in this study and the amplicon size respectively was 839 bp and 970 bp. The RT-PCR technique has been successfully used by numerous researchers (Sajo et al., 2022 ![]() ; Mehmood et al., 2019
; Mehmood et al., 2019 ![]() ; Shofa et al., 2018
; Shofa et al., 2018 ![]() ; Ewies et al., 2017
; Ewies et al., 2017 ![]() ). Matrix gene detection of NDV was also obtained by Ak et al. (2019
). Matrix gene detection of NDV was also obtained by Ak et al. (2019 ![]() ); and Shah et al. (2019
); and Shah et al. (2019 ![]() ). Utilizing the RT-PCR technique along with sequencing to classify NDV field isolates into velogenic, mesogenic, and lentogenic strains will provide valuable insights into the types of strains circulating in a specific geographical area. This approach will help gather epidemiological data regarding changes in antigenicity and pathogenicity of field isolates. Furthermore, it will aid in developing vaccination strategies aimed at effectively controlling the disease in field conditions.
). Utilizing the RT-PCR technique along with sequencing to classify NDV field isolates into velogenic, mesogenic, and lentogenic strains will provide valuable insights into the types of strains circulating in a specific geographical area. This approach will help gather epidemiological data regarding changes in antigenicity and pathogenicity of field isolates. Furthermore, it will aid in developing vaccination strategies aimed at effectively controlling the disease in field conditions.
4.3 Pathotypical characterization of NDV
Pathotyping of ND viruses hinges on their virulence rather than antigenic differences. NDV strains are classified as velogenic (highly virulent), mesogenic (moderately virulent), or lentogenic (low virulent) based on specific pathogenicity indices. Velogenic strains typically have an MDT of less than 60 hours, an ICPI greater than 1.4, and an IVPI ranging from 2 to 3. Mesogenic strains exhibit an MDT of 60-90 hours, an ICPI between 1 and 1.4, and an IVPI around 0.5. Lentogenic strains show an MDT over 90 hours, an ICPI of 0 to 0.2, and an IVPI of 0. These indices are essential for categorizing NDV strains by their pathogenic characteristics, crucial for managing outbreaks in poultry populations. In the current pathotypical characterization investigation, the MDT and ICPI values were 54.34 and 1.55, respectively. MDT of less than 60 hours and ICPI of greater than 1.5 meet all requirements for virulent strains. All isolates induced the demise of inoculated embryonated chicken eggs after 72 hours post-inoculation. Based on MDT and ICPI for one isolate, our result indicated that all the local samples had virulent NDV strains (velogenic). These pathogenicity tests were performed by some workers Susta et al. (2011 ![]()
5. Conclusions
This study successfully identified four positive isolates from post-mortem samples of suspected birds. Initial screening through slide hemagglutination tests and subsequent confirmation via RT-PCR demonstrated the presence of Newcastle disease virus. The use of fusion and matrix gene-specific primers in RT-PCR, followed by passage into chicken embryonated eggs, allowed for detailed observation and analysis. The slide HA test provided primary identification, while further molecular confirmation was achieved through RNA extraction and conversion to cDNA. This comprehensive approach validated the status of the Newcastle disease virus in all isolates, with one field isolate classified as velogenic based on pathotypical analysis. The findings underscore the effectiveness of the applied methodologies in diagnosing and characterizing virulent strains of Newcastle disease virus, meeting all necessary criteria for virulence.
Acknowledgements
The research was funded by the Bangladesh Agricultural University Research System (BAURES), Bangladesh Agricultural University, Mymensingh-2202, Bangladesh. Project title: Pathotypical and genotypical characterization of strains of Newcastle Disease virus isolated from outbreak in chickens flocks in some regions of Bangladesh. Project No. : 2021/62/BAU.
Ethical statement
Not applicable.
Data availability statement
The data generated from this study might be available on the valid request from the corresponding author.
Informed consent statement
No informed consent was required to conduct the study.
Conflict of interest
The authors declare no conflict of interest.
Author contributions
Conceptualization: Limon Biswas and Mohammad Ferdousur Rahman Khan; Data collection: Limon Biswas and Dula chakraborty; Data analysis: Limon Biswas and Najmun Nahar Popy; Conducted data analysis and interpretation of results: Limon Biswas and Mithun Talukder; Designed the experiment and edited the manuscript: Limon Biswas and Mohammad Habibur Rahman; Figure preparation: Limon Biswas and Md. Bahanur Rahman; Reviewed it and revised the final version. All authors critically reviewed the manuscript and agreed to submit final version of the article.



